In late 2019, a new infectious respiratory disease, COVID-19, emerged in Wuhan, China. After spreading to hundreds of countries worldwide, COVID-19 is now set to be the largest global pandemic since the 1918/1919 “Spanish flu”. While over a century has now past, certain measures to combat infectious disease remain similar – some countries are in lockdown, social distancing is practised and mask-wearing is encouraged. However, our scientific knowledge and the corresponding technology has also rapidly advanced in the last century. One particular example is our understanding of nucleic acids and their genetic code.
DNA (double-stranded) or RNA (single-stranded) are both polynucleotides linked by a phosphate backbone; each nucleotide is formed of a nitrogenous base (or “letter”), a pentose sugar and a phosphate group. The four “letters” are adenine (A), guanine (G), cytosine (C), and thymine/uracil (T/ U, depending if the nucleic acid is DNA/RNA respectively). As published by Watson and Crick in 1953[1], these bases can form a specific pattern by hydrogen-bonding, in which G “pairs” with C, and A “pairs” with T. This base-pair recognition is essential both for reproduction of cells (wherein the genetic code is copied), and protein synthesis (wherein the genetic code is translated into an amino acid sequence, the building blocks of proteins).
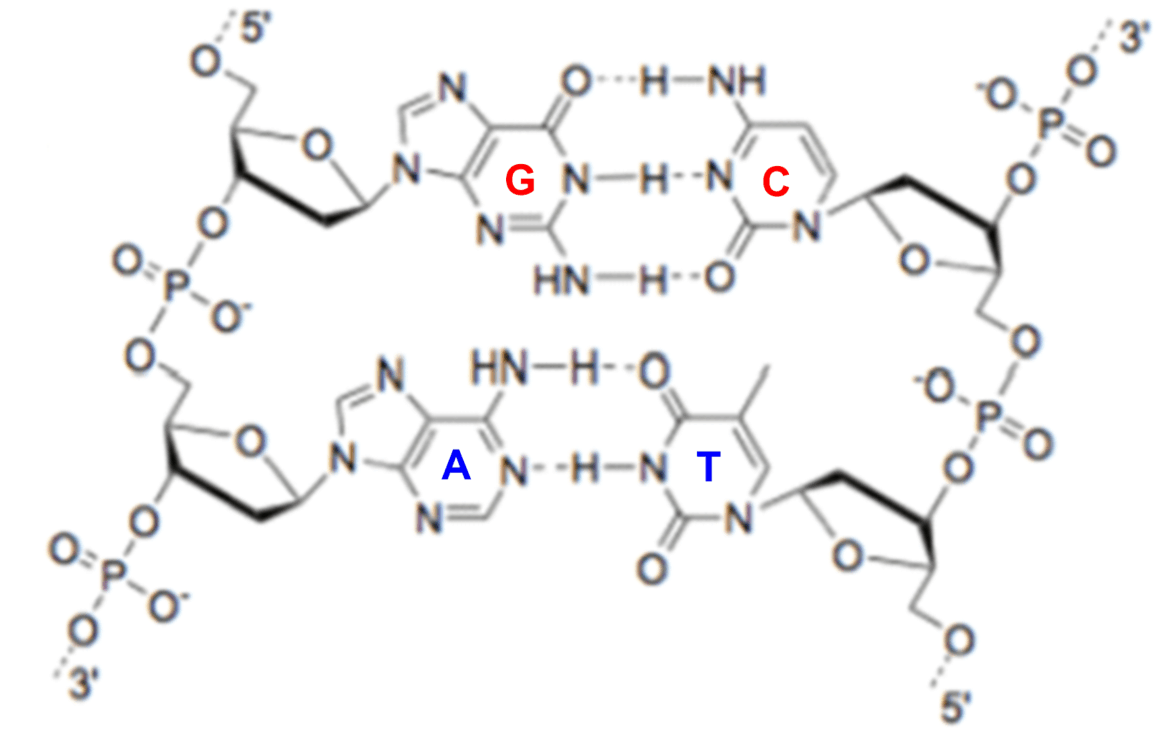
A representation of base-pairing in DNA:G base-pairs with C and A base-pairs with T via hydrogen bonding (see dashed lines).
Since 1953, the technology has expanded further such that scientists can now readily sequence and synthesize nucleic acid building blocks at increasingly lower costs.[2]
This technology has now been utilized against COVID-19. By February 2020, the RNA sequence of the novel coronavirus SARS-CoV-2 had already been deduced.[3] [4] From clinical samples, viral RNA was extracted, quantified, transcribed into DNA, then sequenced using next-generation sequencing. Here, we describe the technology involved in each step:
Nucleic acid (RNA) extraction
Nucleic acids must first be separated from other biological molecules in the clinical sample before they are processed further. While RNA can be purified from the biological sample via several methods, one of the first patient samples of SARS-CoV-2 was extracted using spin-column nucleic acid purification[3].
US7267950 describes this type of technology. A biological sample is applied to a material (e.g. modified silica gel column) in the presence of chaotopic agents and salts under negative pressure. The material can selectively bind nucleic acids, while other biological molecules (e.g. proteins) pass through, the column aided by centrifugation. After washing steps, purified RNA can finally be eluted (e.g. using water).
Concentration of nucleic acid (RNA)
Next, on the way to ultimately sequencing the viral RNA, the quantity of extracted RNA was determined fluorometrically[3]. EP2610315 and WO 00/66664 both describe examples of substituted cyanine dyes used to “stain” nucleic acids. These cyanine molecules both show a significant fluorescent enhancement when associated with nucleic acids as compared to when not associated. The fluorescent signal can be detected by a fluorimeter to quantify the amount of RNA in the sample.
RNA reverse transcription
For sequencing, the RNA must then be converted into DNA by reverse transcription. Using natural base pairing (as shown above), reverse transcriptase enzymes can be used to generate a complementary DNA (cDNA) strand from the RNA sequence. Such technology is utilised, for example, in EP0871780. In this patent, a template switching oligonucleotide is utilised to permit template-based extension of the cDNA.
Library preparation and next-generation sequencing
A sequencing “library” must then be prepared. This involves adding DNA adaptors to the nucleic acid sample with a DNA ligase enzyme (“a DNA glue”) such that the DNA is compatible with NGS instruments (one example of this type of technology is described in WO201534552). The polymerase chain reaction (PCR) is then used to generate enough DNA for sequencing. PCR, as described in US4683195, uses heat-sensitive polymerase enzymes (enzymes used in nature to replicate DNA via base-pairing) to “copy” strands of the DNA template.
The viral genetic code can then be sequenced. Next-generation DNA sequencing technology uses a “sequencing by synthesis approach” as described in WO 03/048387 . The DNA sequence is determined stepwise by fluorescent nucleotides that are inserted against a template strand using a DNA polymerase. Each nucleotide is labelled with a specific fluorophore to distinguish each base. This gives a detectable read-out of the sequence, since G pairs with C, and A pairs with T. A protecting group on the fluorescently-labelled nucleotide temporarily stalls polymerization such that the incorporation event can be detected. This protecting group is then removed and this process can then be repeated along the DNA strand.
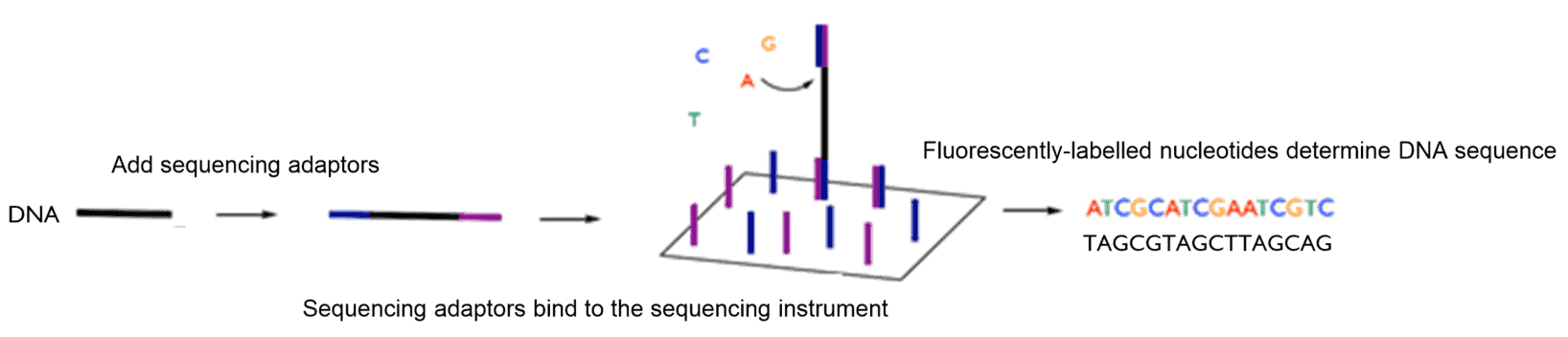
While determination of the genetic code is just one of many breakthroughs in our fight against COVID-19, this has enabled the scientific community and governmental organisations to at least:
- More reliably diagnose patients with COVID-19 and track and trace those that have come into contact with a confirmed case[5]; Clinical samples from patients can be tested to see if they contain any SARS-CoV-2 RNA. Individuals that have come into contact with a “confirmed” COVID-19 case can be tested for infection. This strategy can be used to detect even asymptomatic cases to slow down the spread of the virus. This can be achieved using PCR methods as described above, where any “copies” of the SARS-CoV-2 sequence can be detected.
- Determine the likely origin of the virus[4]SARS-Cov-2 shares a significant sequence similarity with coronaviruses found in bats. This may lead to the implementation of certain regulations to prevent similar viruses emerging in the future.
- Study the global transmission and mutation rate of the virus[6]; Over 5000 viral genomes have been sequenced around the world by early May 2020. By continually sequencing the genome, scientists can monitor how the virus changes and spreads around the world.
- Determine the structure of viral proteins[7]SARS-CoV-2 proteins have now been expressed or modelled by many research groups using the viral genetic code. This is crucial for understanding how the virus interacts with the human host during the course of infection; enabling scientists to identifying potential targets for therapeutic intervention. The viral “spike proteins”, for example, have already been used for the development of vaccines against the virus.[8][9]
We are proud of all researchers currently working to tackle this pandemic, and thank all scientists (past and present) for their contribution. Here at Haseltine Lake Kempner, everyone affected by COVID-19 continues to be in our thoughts and we hope you are all keeping safe.
[1] Watson, J., Crick, F. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 171, 737–738 (1953).
[2] https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data
[3] Wu, F., Zhao, S., Yu, B. et al.A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
[4] Zhou, P., Yang, X., Wang, X. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020)
[5] https://www.england.nhs.uk/coronavirus/wp-content/uploads/sites/52/2020/03/guidance-and-sop-covid-19-virus-testing-in-nhs-laboratories-v1.pdf
[6] https://nextstrain.org/ncov
[7] Gordon, D.E., Jang, G.M., Bouhaddou, M. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature (2020).
[8] Grant, O.C, Montgomery, D. Ito, K., Woods, R.J. 3D Models of glycosylated SARS-CoV-2 spike protein suggest challenges and opportunities for vaccine development, bioRxiv 2020.04.07.030445
[9] http://www.ox.ac.uk/news/2020-04-23-oxford-covid-19-vaccine-begins-human-trial-stage